Reviewed manuscript: Mühlfeld, C., J.R. Nyengaard, and T.M. Mayhew (2010) A review of state-of-the-art stereology for better quantitative 3D morphology in cardiac research. Cardiovascular Pathology, 19, pp. 65 – 82.
The aim of this review is to encourage those doing cardiac research to use unbiased stereology as is done in other fields such as Pulmonary, Kidney, Placenta, and Neuroscience (Mühlfeld, 2010, third paragraph). The use of thin sections is recommended to ‘improve resolution and allow us to visualize internal structural detail’ (Mühlfeld, 2.1, first paragraph, second sentence). However, recently developed histological techniques can allow for good visualization with thick sections.
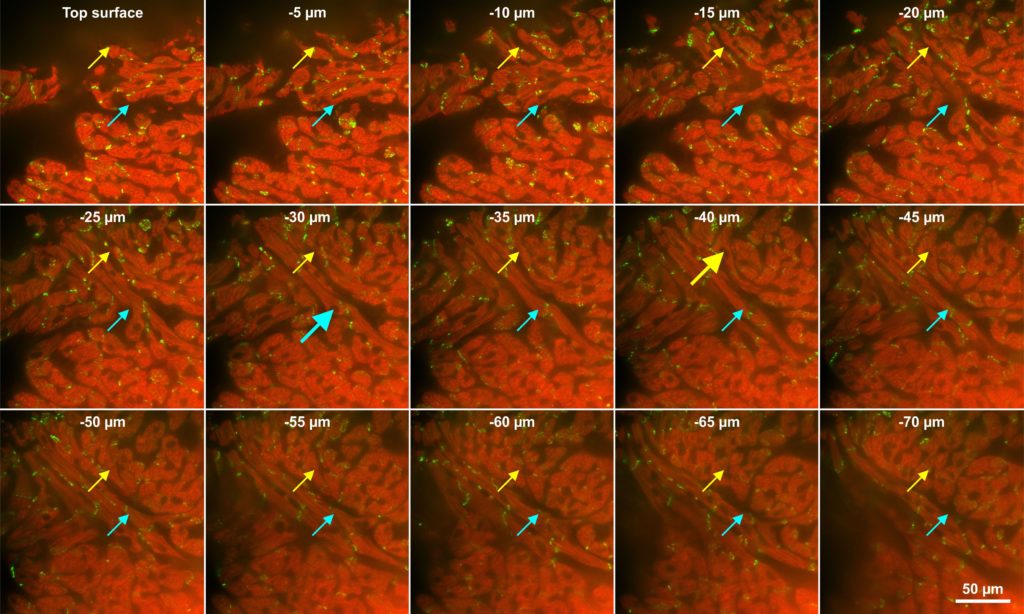
Eighty micron thick section from mouse heart. Section was processes with CLARITY, followed by immunofluorescence against troponin (red) and connexin 43 (yellow), and imaged with an Olympus DSU microscope. The two different colored arrows follow two individual cardiac myocytes through the z-axis, large arrows indicate a cross section through the nucleus. Image courtesy by Dr. Eckhard U. Alt, Heart and Vascular Institute, Department of Medicine, Tulane University, New Orleans, LA, USA
The suggestions given in this review for unbiased stereology on thin sections of cardiac tissue are valid; but we will also include suggestions for using thick sections, like in the image above, because probes that can be used on thick sections do not require isotropic or vertical sections.
The authors characterize the use of unbiased stereological methods in biomedical research as aiming to ‘obtain quantitative information about three-dimensional (3D) features of tissues, cells, or organelles from two-dimensional physical or optical sections’. Examples of important questions are given: ‘Is there a significant loss of cardiomyocytes during progression from ventricular hypertrophy to heart failure?’ and ‘Does a specific treatment reduce the degree of fibrosis in the heart?’. But it is also pointed out that the use of unbiased stereology is rare for cardiac research (Mühlfeld, abstract). Therefore the goal of this review is to make clear how to estimate ‘hard, biologically useful information’ regarding number, length, surface, or volume of structures within a well-defined region –in a way that ‘should be simple and provide a precise estimate of the real value at low cost’ in other words, efficiently; by encouraging the use of design based stereology; described as the ‘gold standard in quantitative microscopy’ (Mühlfeld, Current status, second paragraph).
The need to understand sources of bias in quantitative 3D microscopy is stressed (Mühlfeld, Table 1). No assumptions about shape, size, or distribution of structures can be used (see design based stereology). No orientation in 3-D space between the probe and the tissue being probed can be favored. This means for thin sections, a process to ensure that the tissue itself is isotropic must be employed, and it is recommended to use the ‘isector’ or ‘orientator’ to ‘spin’ tissue blocks to make their contents isotropic (see isotropic and vertical sections). A cautionary example is given involving the determination of the surface area of papillary muscles in cross-section. If papillary muscles are only sectioned and visualized in cross section, the surface area will be underestimated because the probe (consisting of lines, e.g. see Merz probe) will make less intersections with the muscle surface than if the probe tissue interaction were isotropic in 3-D space. Another example given is estimating the length of capillaries in the heart (Mühlfeld. 2.2). It is pointed out that ‘the orientation of the capillaries in cardiac muscle is related to the orientation of the cardio myocytes’ (Mühlfeld, 2.3, first paragraph, first sentence). If you use a probe that is a surface and count the intersections with the vessels (see using image as probe for length), and vertical or isotropic sections are not used, you are in danger of over- or under-estimating the surface area. For example if the cardiac myocytes are sectioned in such a way that longitudinal orientation is favored, the capillaries will also be mostly longitudinal, and their surface will be over-estimated because more intersections are counted than if the interaction between probe (line) and tissue (surface) was isotropic. One solution is to manipulate the tissue blocks before thin-sectioning so that cross-sections will not be favored; there should be an equal chance to get any section, cross-, longitudinal-, or all possible orthogonal-sections. We however, recommend the use of thick sections so that thick section probes can be used and the tissue can be sectioned as the researcher wants to section (see preferential sections). Another source of bias mentioned is tissue shrinkage (Mühlfeld, 2.5 Tissue Shrinkage), the suggestion is to be aware of the properties of the embedding medium and measure pre- and post-shrinkage thicknesses, especially for volume, surface and length. It is mentioned that the optical fractionator to estimate number, is not affected by shrinkage artifact. The next source of bias discussed is the ‘volume reference trap’ that is a danger when using the ‘numerical density multiplied by reference volume’ (NvVref) method. The authors suggest being careful about measuring the reference volume. Possible techniques for measuring the Vref are given as water displacement, weighing and dividing by muscle density, or point-counting. It is said that a reasonable precision of the volume estimate is obtained by counting 100 – 200 points on 7 – 12 sections in total per organ (Mühlfeld, 2.4) and Howard and Reed is listed as the source. A good example of the volume reference trap is given involving the number of cardiac myocytes per volume (Mühlfeld, Fig. 4). If only densities are reported readers will not know if for instance, an increase in density is caused by more myocytes or less volume or some combination. It is lamented that ‘Unfortunately, the literature is littered with articles reporting quantities as densities in 2D or 3D but interpreting them as if they were absolute quantities’ (Mühlfeld, 2.4, second to last paragraph, second sentence and see Table 4). We suggest using fractionator probes instead of NvVref probes to avoid this problem. The requirement for systematic random sampling to avoid bias is stressed (Mühlfeld, Fig. 1 and 2). Also pointed out is the need to use sections during point-counting that are not a large proportion of the whole region; one tenth of the whole region at most, to avoid ‘over-projection’ effects that can cause bias. Finally a warning is given against using probes that will make volume-weighted estimates of particle-volume (e.g. see Point Sampled Intercepts), if number-weighted estimates of particle-volume are what is needed (e.g. see Nucleator with Disector).
In the section of the this review called ‘Estimating Myocardial Structures’ the authors give recommendations for unbiased stereology probes for thin sections to be used with the NvVref method (see table 3 and 4). We however, recommend using thick section probes that use the fractionator method, when possible. Consider a cornerstone of Cardiac research with implications for development and disease: determining the number of cardiac myocytes in the heart; “Probably one of the most valuable stereological parameters in the heart is cardiomyocyte number’ (Mühlfeld, pg. 74, third paragraph, first sentence). Let’s first look at some examples where unbiased stereology was not used. In the conclusion of the Mühlfeld review, it is stated that they are not trying to depreciate former morphological results but instead to encourage unbiased stereology in the future. We agree and would like to look at these examples in the same light. In one study on cardiomyocyte endowment at birth, a ‘temporary bubble in DNA’ was used to attempt to measure the number of cardiac myocytes (Thornburgh, 2011, first paragraph). In another study looking at how systolic pressure load may change the number of cardiomyocytes, point-counting was used to estimate the percent by volume of cardiomyocytes in the heart (Barbera, 2000). They then divided that by the volume of each individual myocyte, although it is not said how the individual volumes are measured. This is a model based process subject to bias since an assumption must be made about the volume of each myocyte. When trying to determine the number of cardiomyocytes in another study (Banerjee, 2007), they looked at a single optical plane at low magnification and set a threshold using Metamorph. This breaks the important rule in unbiased stereology that is pointed out in the Mühlfeld review (Mühlfeld, 2.2) ‘the information about a zero-dimensional structural feature, namely, number, is not present in one 2D section’. Also the use of a threshold by a computer program is not well integrated into any stereological techniques yet (Schmitz, 2014). More examples of techniques other than unbiased stereology are given in Table 4 of the Mühlfeld review.
The best way to estimate the number of particles (in this case cardiac myocytes), rather than techniques like those described above, is to use the optical fractionator for thick sections, or if you can’t obtain or use thick sections, the physical fractionator for thin sections. In the Mühlfeld review, it is noted that the optical fractionator is better for finding the leading edges of complex objects, and pointed out also that the connectivity assay could be used to account for complex cell tops where it is hard to determine the leading edge. They call this ‘resorting’ to the use of the optical fractionator, probably due to the fact that at the time visualization of the cardiac myocytes was not as good in the thick sections required for the optical fractionator. If thick sections can be used (see image at top) the optical fractionator is easier to use, although computer programs that help match reference- to lookup-sections make physical fractionator easier than it was (Mühlfeld , Fig. 5, legend, last sentence). Both the optical- and physical-fractionator probes use the disector and its counting rules to avoid over-counting the number of cells. This review sites Sterio’s 1984 paper for the disector and states that ‘stereology entered a different era’ … ‘for the design-based quantification of number and size of particles’ (Mühlfeld, 3.2, first sentence). One of the disector rules is that the leading edge of the particle, or some unique point such as the nucleolus, if there is only one per cell, must be visualized and then used to count the cell; if this unique point is captured by the disector, the whole cell is counted. A big problem in using the optical fractionator on cardiac myocytes is that it is very hard for a human to find their leading edges (see image above). That is why in the past, when unbiased stereology has been used to estimate the number of cardiomyocytes, the nuclei of the fibers are used; the leading edge of a cardiac myocyte nucleus is much easier to spot and focus-on than the leading edge of the whole cell. In their study (Austin, 1995), the authors did just this because this approach that estimates number, not density, is preferable to ‘those which are limited to the packing densities of nuclei or their sectional images on tissue sections’ (Austin, 1995, third paragraph). They use the physical disector to estimate number of myocytes in muscle layer (myocardium) of human heart. They wanted to establish a protocol to look at patterns of cardiac growth using archival material. Two control and four ‘abnormal’ human hearts were studied. Heart ventricles were removed of clots, fixed, and weighed, and the ventricles cut into small pieces and weighed. Several pieces were then picked randomly for resin embedding. Four to five contiguous thin sections were taken on a microtome at 3 microns and stained with hematoxylin and eosin; the interval of the sections is not given. Three microns is less than the smallest nucleus so it won’t ‘miss’ nuclei when comparing two contiguous sections. Systematic random sampling was used (SRS), the area-sampling-fraction is not given but they did say the mean number of counting frames is fifteen. The authors made slides and double-projected images of the reference and lookup sections next to each other on a screen (stiff white card) and the counting frames (1,217 square microns) were present on the screens (for a modern example please see minute 28:30 of this movie). They counted myocyte nuclei using disector counting rules: if the cross section of a nucleus is present in the reference but not the lookup image, the top of the nucleus has been found, and the nucleus is counted. The NvVref method was used. This means the number per volume is estimated by taking the number of nuclei tops detected and dividing by the volume sampled, where volume sampled is the area of a counting frame times the tissue thickness. The tissue thickness was measured post-artifact, however, after tissue shrinkage had occurred. That gave them Nv. Then they arrived at Vref from the weight of the heart times the assumed density of heart. Nv times Vref gives the absolute number estimate of cardiac myocyte nuclei for the whole heart, and this was multiplied by the fraction of actual myocardium in the ventricle pieces estimated with point counting (Area Fraction Fractionator). The parameters were not given for the point-counting. They recognize the post-artifact thickness used to calculate the volume to be used for Nv is not appropriate to use with the pre-artifact Vref, but say it will vary with both groups equally. This is an assumption that is hard to prove and ideally never used in an unbiased stereology study. It may have been better to use the pre-shrinkage-artifact thickness to calculate the volume for Nv. We recommend avoiding the NvVref method and instead using the Fractionator method to avoid these problems that come along with calculating an Nv and measuring a Vref. With the Fractionator method there is no need for Vref. Lastly they note that they are not counting cells but instead their nuclei because there can be more than one nucleus per cell.
As discussed above, a major pitfall to estimating the number of cardiomyocytes using disector counting rules during the optical or physical fractionator, is that it is difficult or impossible for a human to easily find their leading edges. Therefore, the leading edge of the nucleus, rather than of the fiber itself, has been used. This leaves the problem of how to know how many cardio myocytes there are. If all of these fibers had one nucleus it would be easy to convert, but ‘Counting the number of myocyte nuclei in the heart is not an unbiased estimate of cardiomyocyte number due to the varying number of nuclei per cardiomyocyte’ (Mühlfeld, 2010, 3.2, second paragraph, second to last sentence). To figure out how many nuclei there are per cardiac myocyte, the idea of counting them per cell in ‘vertical windows’ has been used with the physical fractionator (Brüell, 2005, Brüell, 2007, Schipke, 2014) and the optical fractionator (Bergmann, 2015). Isotropic uniform random sections are used, and when a myocyte is sectioned longitudinally, it is followed all the way through and the number of nuclei are counted (Schipke, 2014, Fig. 1).
Suggestions for unbiased stereological probes to be used on thin sections to estimate length, surface, and volume of structures in the heart are also given (Mühlfeld, section 3). Using the image plane as the probe to estimate length is recommended for thin sections, but to avoid the need for vertical or isotropic tissue sectioning, we recommend using thick sections if possible and using the Spaceballs probe. For estimating surface on thin sections a test-line grid is suggested for thin sections, but if one would like to section the tissue preferentially, we suggest obtaining thick sections and the Isotropic Fakir probe. When it comes to estimating the volume of regions, the Cavalieri-point-counting probe is actually designed for thin sections, whether they are physical or optical. Regarding estimating the volume of a cardiac myocyte, point counting is suggested, but due to problems in identifying the cell borders, the Nucleator probe could also be considered (Bergmann, 2015). The nucleator probe requires thick, isotropic or vertical sections, and must be used with the optical disector for a number weighted estimate of volume.
This review (Mühlfeld, 2010) is an excellent guide to thin-section, unbiased, NvVref, stereology. However if thick sections can be used, it will obviate the need to manipulate the tissue blocks for vertical or isotropic sectioning to assure isotropy of interaction between the probe and the tissue. Furthermore, if the volume fraction can be kept track of, the use of fractionator probes instead of NvVref probes would also greatly simplify the process.
Austin A1, Fagan DG, Mayhew TM. (1995) A stereological method for estimating the total number of ventricular myocyte nuclei in fetal and postnatal hearts. J Anat. 1995 Dec;187 (Pt 3):641-7.
Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, Sjostrom SL, Szewczykowska M, Jackowska T, Dos Remedios C, Malm T, Andrä M, Jashari R, Nyengaard JR, Possnert G, Jovinge S, Druid H, Frisén J. (2015) Dynamics of Cell Generation and Turnover in the Human Heart. Cell. 2015 Jun 18;161(7):1566-75.
Brüell, A and J.R. Nyengaard (2005) Design-based stereological estimation of the total number of cardiac myocytes in histological sections. Basic Res. Cardiol. 100: 311-319.
Brüell, A, Christoffersen, E.H. and J.R. Nyengaard (2007) Growth hormone increases the proliferation of existing cardiac myocytes and the total number of cardiac myocytes in the rat heart. Cardiovascular Research 76, 400-408.
Banerjee I, Fuseler JW, Price RL, Borg TK, Baudino TA. (2007) Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am J Physiol Heart Circ Physiol. Sep;293(3):H1883-91.
Barbera A, Giraud GD, Reller MD, Maylie J, Morton MJ, Thornburg KL. (2000) Right ventricular systolic pressure load alters myocyte maturation in fetal sheep. Am J Physiol Regul Integr Comp Physiol. Oct;279(4):R1157-64.
Sterio DC. (1984) The unbiased estimation of number and sizes of arbitrary particles using the disector. J Microsc. 1984 May;134(Pt 2):127-36.
Howard and Reed (2010) Unbiased Stereology, 2nd edition, QTP Publications, Coleraine. UK.
Mühlfeld, C., J.R. Nyengaard, and T.M. Mayhew (2010) A review of state-of-the-art stereology for better quantitative 3D morphology in cardiac research. Cardiovascular Pathology, 19, pp. 65 – 82.
Schipke J, Banmann E, Nikam S, Voswinckel R, Kohlstedt K, Loot AE, Fleming I, Mühlfeld C. (2014) The number of cardiac myocytes in the hypertrophic and hypotrophic left ventricle of the obese and calorie-restricted mouse heart. J Anat. Nov;225(5):539-47.
Schmitz C, Eastwood BS, Tappan SJ, Glaser JR, Peterson DA, Hof PR. (2014) Current automated 3D cell detection methods are not a suitable replacement for manual stereologic cell counting. Front Neuroanat. 2014 May 7;8:27.
Thornburg K1, Jonker S, O’Tierney P, Chattergoon N, Louey S, Faber J, Giraud G. (2010) Regulation of the cardiomyocyte population in the developing heart. Prog Biophys Mol Biol. 2011 Jul;106(1):289-99.